在单细胞研究逐步进入规模化的时代,你是否还在担心单细胞研究“门槛”过高?
为了满足不同客户的科研需求,降低单细胞研究“门槛”,吉因加基于国产单细胞DNBelab C4技术平台,特此隆重推出单细胞限时优惠活动,客户可在单细胞分辨率下从细胞异质性、细胞功能、细胞互作及基因网络调控等多维度解锁生命的奥秘。
国产DNBelab C4单细胞平台,基于高通量单细胞转录组捕获系统DNBelab C4,搭载国产DNBSEQ超高通量测序系统,以及开源的数据质控软件HT-RNA分析包,可实现高通量单细胞转录组文库测序和数据分析挖掘。该平台结合吉因加自身在单细胞技术研发方面的创新沉淀和超高通量测序大平台,匹配严格的质控体系和特色吉云交互式数据挖掘系统,旨在为广大用户提供高通量、样本兼容性强、性价比高的一站式单细胞组学研究的全流程测序服务,为人和动植物等的单细胞研究提供更加全面、精准的服务。
国产平台礼遇重磅福利 实现高性价比
国产DNBelabC4单细胞捕获系统,精准匹配吉因加日产54T超高通量DNBseq-T7测序大平台,产生高质量数据同时降低了成本,性价比高。

吉因加实测样本指标表现优秀
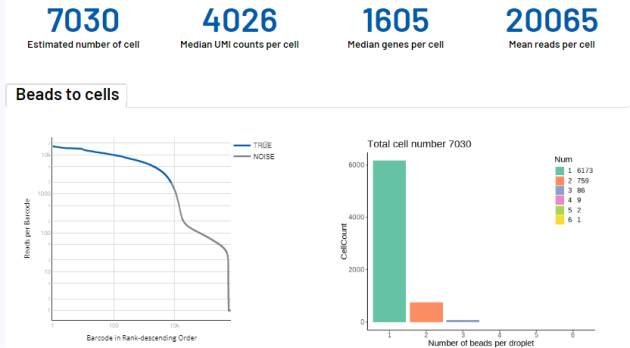
吉因加部分单细胞实测数据的展示(人组织类型)
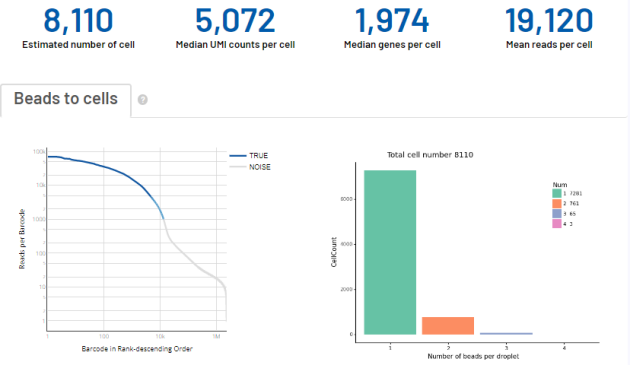
吉因加部分单细胞实测数据的展示(小鼠组织类型)
丰富的项目经验
目前DNBelabC4平台已完成130+例样本检测服务,覆盖人、小鼠、动物等多物种,兼容组织解离、抽核2种处理方案,处理组织类型超40+,涉及细胞系、PBMC、血液、肝、 肺、脾、胸腺、子宫内膜、淋巴结、卵巢穿刺组织、胃癌、肝癌、胸腺瘤、口腔癌、宫颈癌、附睾等多组织类型。
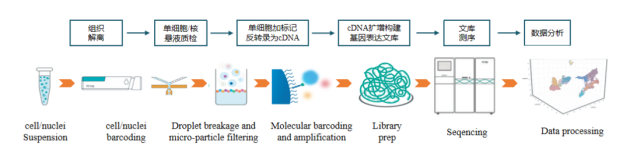
吉因加全流程服务
依托DNBelab C4系统,吉因加为客户提供从方案指导→细胞悬液制备→单细胞分离→文库建库→测序→数据分析→数据挖掘一站式服务。
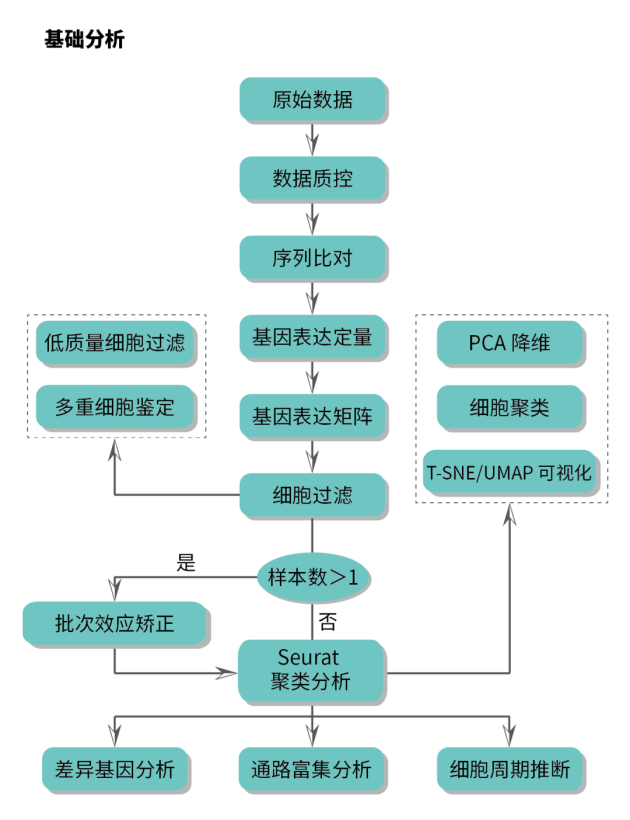
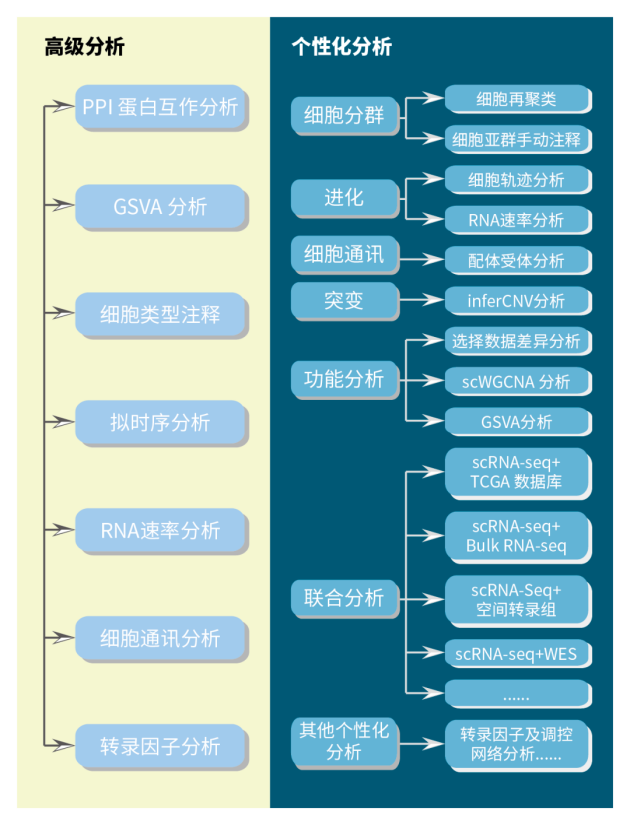
专业+全面的分析内容
吉因加强大的单细胞科研团队和吉云平台的支撑下,不仅可以提供基础+高级+个性化的全面分析内容,而且支持客户在线使用吉云平台流程及分析工具自主进行数据挖掘。
16项标准分析大礼包,可满足各类科研客户实际分析需求,全面助力单细胞数据分析
10项个性化分析内容助力深度挖掘单细胞数据
DNBelab C4助力多篇高分文章
DNBelab C系列单细胞分选平台已助力发表文章23篇,平均影响因子多达17+。其中10分以上高分文章多达8篇,2篇文章发表在国际顶级学术期刊Nature上。
序号发表时间期刊影响因子文章题目12022/4/13Nature69.504Cell transcriptomic atlas of the adult non-human primate Macaca fascicularis22022/3/22Nature69.504Rolling back of human pluripotent stem cells toan 8-cell embryo-like stage32022/3/10Cell Research46.297Evolutionarily conservative and non-conservative regulatory networks during primate interneuron development revealed bysingle-cell RNA and ATAC sequencing42020/7/19Immunity43.474Single-Cell Sequencing of PeripheralMononuclear Cells Reveals Distinct Immune Response Landscapes of COVID-19 andInfluenza Patients52022/6/16Nature Ecology&Evolution19.1A single-cell transcriptomic atlas tracking the neural basis of division of labor in an antsuperorganism62022/6/24Nature Communications17.694Endothelial cell heterogeneity and microglia regulons revealed by a pig cell landscape atsingle-cell level72022/5/4Developmental Cell13.417Spatiotemporal mapping of gene expression landscapes and developmental trajectoriesduring zebrafish embryogenesis82021/3/5Theranostics11.6Single-cell transcriptome analysis of the heterogeneous effects of differential expressionof tumor PD-L1 on responding TCR-T cells92021/1/1Clnicaland TranslationalMedicine8.554Multiregion single-cell sequencing reveals the transcriptional landscape of the immunemicroenvironment of colorectal cancer102022/1/29Clinicaland TranslationalMedicine8.554Molecular mechanisms governing circulating immune cell heterogeneity across differentspecies revealed by single-cell sequencing112022/5/6Clinical and TranslationalMedicine8.554Single-cell atlas of peripheral bloodmononuclear cells from pregnant women122022/6/14Scientifc Data8.501A map of bat virus receptors derived from single-cell multiomics132021/12/1Analytical Chemisty8.008High-Throughput Functional Screening of Antigen-Specific T Cells Based on DropletMicrofluidics at a Single-Cell Level142022/5/23Analytical Chemistry8.008Generation and Screening of Antigen-SpecificNanobodies from Mammalian Cells Expressing the BCR Repertoire Library Using Droplet-BasedMicrofluidics152021/5/17Frontiers in molecularneuroscience6.261Single-Nucleus Chromatin Accessibility Landscape Reveals Diversity in RegulatoryRegions Across Distinct Adult Rat Cortex162022/2/2Iscience6.107Single-cell multiomics reveals heterogeneous cell states linked to metastatic potential in livercancer cell lines172022/3/1Front. Cell Dev.Biol.6.081Single-cCelIAtlasof the Chinese Tongue Sole (Cynoglossus Semilaevis)Ovary RevealsTranscriptional Programs of Oogenesis in Fish182022/4/5Front. Cell Dev.Biol.6.081Integrative single-cell RNA-seq and ATAC-seq analysis of mesenchymal stem/stromal cellsderived from human placenta192022/4/6Front. CellDey. Biol.6.081Transcriptomic profile of the mouse postnatalliver development by single nucleus RNA sequencing202021/5/20Journal of Genetics andGenomics5.723Ahigh-resolution cell tlas of the domestic pig lung and an online platform for exploring lungsingle-cell data212022/1/22Journal of InflammationResearch4.631Single Nuclear RNA Sequencing Highlights Intra-Tumoral Heterogeneity and Tumor Microenvironment Complexity in TesticularEmbryonic Rhabdomyosarcoma222021/4/2Biochemical and BiophysicalResearch Communications3.322Single-cell transcriptional diversity of neonatal umbilical cord blood immune cells reveals neonatal immune tolerance232022/1/21eInsightSingle-Cell Transcriptomic Landscape Identifies the Expansion of Peripheral Blood Monocytes asan Indicator of HIV-1-TB Co-Infection
郑重声明:图文由自媒体作者发布,我们尊重原作版权,但因数量庞大无法逐一核实,图片与文字所有方如有疑问可与我们联系,核实后我们将予以删除。
热门阅读
最近发表
商业之声,版权所有